A quick introduction to CRISPR
Julian PampelCRISPR stands for "Clustered regularly interspaced short palindromic repeats". They were discovered in prokaryotic DNA (E.coli) first in 1987 (Y. Ishino et. al) and contain short repetitions of base sequences. In the course of better sequencing techniques in the early 2000s similar sequences were found in other bacteria as well. Further research revealed that the sequences of repeats in Bacteria chromosomes is interspaced with sequences derived from viruses. So called Cas proteins (CRISPR-associated genes) encodes putative nuclease or helicase proteins (homology to DNA repair enzymes). Today we know that the CRISPR-System helps bacteria to acquire a type of immune system.
Principle of acquired immune system in bacteria
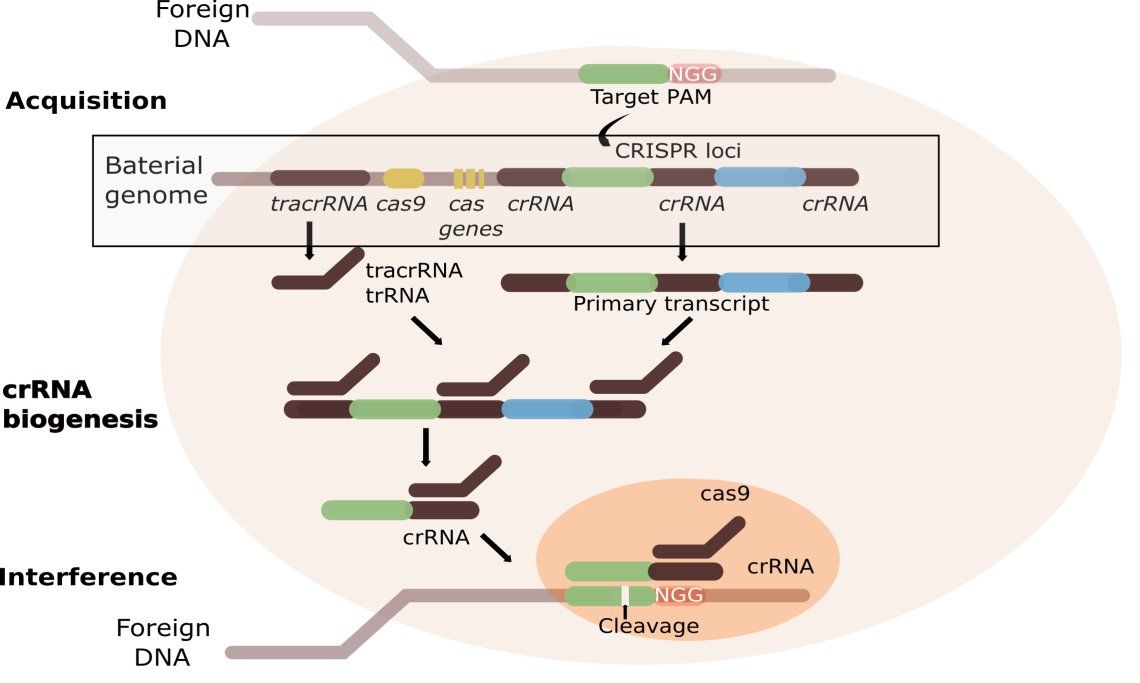
The bacterial cell detects foreign viral DNA with the help of PAM. The protospacer adjacent motif (PAM) is a 2-6 base pair DNA sequence immediately following the DNA sequence targeted by the Cas9 nuclease in the CRISPR bacterial adaptive immune system. PAM is a component of the invading virus or plasmid, but is not a component of the bacterial CRISPR locus. Cas9 will not successfully bind to or cleave the target DNA sequence if it is not followed by the PAM sequence, thereby preventing the CRISPR locus from being targeted and destroyed by nuclease.
CRISPR Plasmids
Off-the-shelf tested plasmids for CRISPR.
Our Cas9 Products
See all of our quality Cas9 enzyme products.
Short pieces of viral DNA (21 to 72 bp ) get integrated in the CRISPR locus as spacer. CRISPR repeats range in size from 24 to 48 base pairs. They usually are palindromic, encouraging a secondary hairpin structure. Repeats are separated by spacers of similar length.
During RNA transcription CRISPR RNAs (crRNAs & tracrRNA) are transcribed from the bacterial genome. TracrRNA is complementary to and base pairs with a pre-crRNA forming an RNA duplex. This is cleaved by RNase III, an RNA-specific ribonuclease, to form a crRNA/tracrRNA hybrid. This hybrid acts as a guide for the endonuclease Cas9, which cleaves the invading nucleic acid and therefore silences the viral gene. A distinguishing feature of CRISPR/Cas systems compared to other sequence specific bacterial defenses, such as restriction modification systems, is that CRISPR/Cas systems are adaptive. Specific Cas proteins recognize foreign genetic elements and integrate their DNA as new spacer sequences into the crRNA array, ultimately allowing the bacteria to adapt and subsequently target these foreign elements
CRISPR in biotechnology
In 2012 Jennifer Doudna and Emmanuelle Charpentier re-engineered Cas9 endonuclease into a more manageable two-component system by fusing the two RNA molecules into a "single-guide RNA" that, when mixed with Cas9, could find and cut the DNA target specified by the guide RNA. They isolated the original Type II CRISPR/Cas System from Streptococcus pyrogenes. By manipulating the nucleotide sequence of the guide RNA, the artificial Cas9 system could be programmed to target any sequence in DNA for cleavage and via vectors in any organism. Providing a DNA repair template allows for the insertion of a specific DNA sequence at an exact location within the genome. The repair template should extend 40 to 90 base pairs beyond the Cas9 induced DNA break. The goal is for the cell's HDR process to utilize the provided repair template and thereby incorporate the new sequence into the genome. Once incorporated, this new sequence is now part of the cell's genetic material and passes into its daughter cells.
Prior to CRISP Zinc finger and TALENS were the method of choice in genomic editing. Zinc finger nucleases which were developed in the early 2000's are synthetic proteins whose DNA-binding domains enable them to create double-stranded breaks in DNA at specific points. In 2010, synthetic nucleases called TALENs provided an easier way to target a double-stranded break to a specific location on the DNA strand. Both zinc finger nucleases and TALENs require the creation of a custom protein for each targeted DNA sequence, which is a more difficult and time-consuming process than that for guide RNAs. CRISPRs are much easier to design because the process requires making only a short RNA sequence. Expressing several gRNAs with the same plasmid ensures that every cell that takes up the plasmid expresses all of the desired gRNAs. Experiments showed it is possible to manipulate up to 62 genes at once, the money as well as time savings are immense. Additionally CRISPR/Cas provides a higher accuracy than both old systems due to less base skipping. This technological advance has fueled efforts not only to edit genomes of a number of different prokaryotic and eukaryotic species, but also as an efficient system for site-specific transcriptional repression or activation.
Plasmid construction and transfer
CRISPR/Cas9 often employs a plasmid to transfect the target cells. When utilized for genome editing, this system includes Cas9, CRISPR RNA (crRNA), trans-activating crRNA (tracrRNA) along with an optional section of DNA repair template that is utilized in either Non-Homologous End Joining (NHEJ) or Homology Directed Repair (HDR). HDR is assumed to be error free because of the use of a template. NHEJ can repair in absence of a template and if the overhangs are perfectly compatible, NHEJ usually repairs the break accurately as well. The repair template is designed for each application, as it must overlap with the sequences on either side of the cut and code for the insertion sequence. The crRNA needs to be designed for each application as this is the sequence that Cas9 uses to identify and directly bind to the cell's DNA. The crRNA must bind only where editing is desired.
| Component | Function |
|---|---|
| crRNA | Contains the guide RNA that locates the correct section of host DNA along with a (hairpin)region that binds to tracrRNA |
| tracrRNA | Binds to crRNA and forms an active complex |
| sgRNA | Single guide RNAs are a combined RNA consisting of a tracrRNA and at least one crRNA |
| Cas9 | Protein whose active form is able to modify DNA (i.e. single strand nicking, double strand break, DNA binding) |
| Repair template | DNA that guides the cellular repair process allowing insertion of a specific DNA sequence |
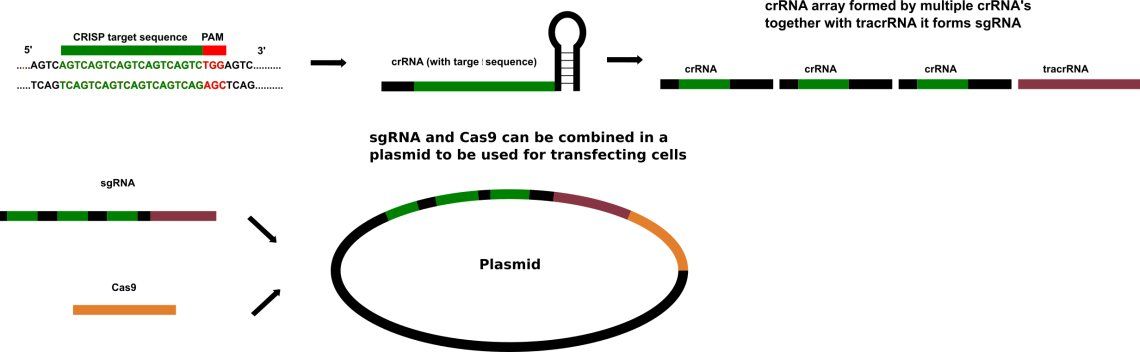
Transfection is the way of choice to deliver Cas9 and gRNA into target cells. Electroporation of DNA, RNA or ribonucleocomplexes is the most common and cheapest system. However, hard-to-transfect cells (stem cells, neurons, hematopoietic cells, etc.) require more efficient delivery systems such as those based on lentivirus (LVs), adenovirus (AdV) and adeno-associated virus (AAV).
CRISPR/Cas9 Products at genomics-online.com
On genomics-online you will find more than 300,000 products for CRISPR/Cas9. Please browse our product portfolio. If you need help with finding the right product, please contact our competent customer support via live chat, email or phone.




